Calculating Climate Anomaly#
Introduction#
A climate anomaly refers to the difference between a measured climate variable and its long-term average or climatology. XArray provides many built-in methods to aggregate and compute statistical averages. In this tutorial, we explore these methods to calculate and visualize soil moisture anomalies.
Overview of the Task#
We will take monthly gridded soil moisture data from NOAA Climate Prediction Center and calculate monthly anomalies relative to the 1971-2000 climatology.
Input Layers:
soilw.mon.mean.v2.nc: A NetCDF file containing monthly mean soil moisture data from 1948 to current.
Output:
Plots of monthly soil moisture anomalies for each month of 2024.
Data Credit:
Global Soil Moisture. NOAA Climate Prediction Center. Retrieved 2025-02
Setup and Data Download#
The following blocks of code will install the required packages and download the datasets to your Colab environment.
%%capture
if 'google.colab' in str(get_ipython()):
!pip install cartopy
import cartopy
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import os
import pandas as pd
import xarray as xr
data_folder = 'data'
output_folder = 'output'
if not os.path.exists(data_folder):
os.mkdir(data_folder)
if not os.path.exists(output_folder):
os.mkdir(output_folder)
Data Pre-Processing#
XArray can directly fetch data from a remote server. We specify the file path to the remote NetCDF file and open it.
filename = 'soilw.mon.mean.v2.nc'
server_url = 'https://psl.noaa.gov/thredds/fileServer/Datasets/cpcsoil/'
data_url = server_url + filename
ds = xr.open_dataset(data_url)
ds
<xarray.Dataset> Size: 959MB
Dimensions: (lat: 360, lon: 720, time: 925)
Coordinates:
* lat (lat) float32 1kB 89.75 89.25 88.75 88.25 ... -88.75 -89.25 -89.75
* lon (lon) float32 3kB 0.25 0.75 1.25 1.75 ... 358.2 358.8 359.2 359.8
* time (time) datetime64[ns] 7kB 1948-01-01 1948-02-01 ... 2025-01-01
Data variables:
soilw (time, lat, lon) float32 959MB ...
Attributes:
Conventions: CF-1.0
title: CPC Soil Moisture
institution: NOAA/ESRL PSD
dataset_title: CPC Soil Moisture
history: Wed Oct 18 15:13:37 2017: ncks -d time,,-2 soilw.mon.mean...
NCO: 4.6.9
References: https://www.psl.noaa.gov/data/gridded/data.cpcsoil.html
source: https://www.cpc.ncep.noaa.gov/products/Soilmst_Monitoring...Select the soilw variable.
da = ds['soilw']
da
<xarray.DataArray 'soilw' (time: 925, lat: 360, lon: 720)> Size: 959MB
[239760000 values with dtype=float32]
Coordinates:
* lat (lat) float32 1kB 89.75 89.25 88.75 88.25 ... -88.75 -89.25 -89.75
* lon (lon) float32 3kB 0.25 0.75 1.25 1.75 ... 358.2 358.8 359.2 359.8
* time (time) datetime64[ns] 7kB 1948-01-01 1948-02-01 ... 2025-01-01
Attributes:
long_name: Model-Calculated Monthly Mean Soil Moisture
units: mm
valid_range: [ 0. 1000.]
dataset: CPC Monthly Soil Moisture
var_desc: Soil Moisture
level_desc: Surface
statistic: Monthly Mean
parent_stat: Other
standard_name: lwe_thickness_of_soil_moisture_content
cell_methods: time: mean (monthly from values)
actual_range: [ 0. 756.0375]We transform the data from 0-360 longitude to -180 - +180 longitudes.
da.coords['lon'] = (da.coords['lon'] + 180) % 360 - 180
da = da.sortby(da.lon)
Calculating Anomaly#
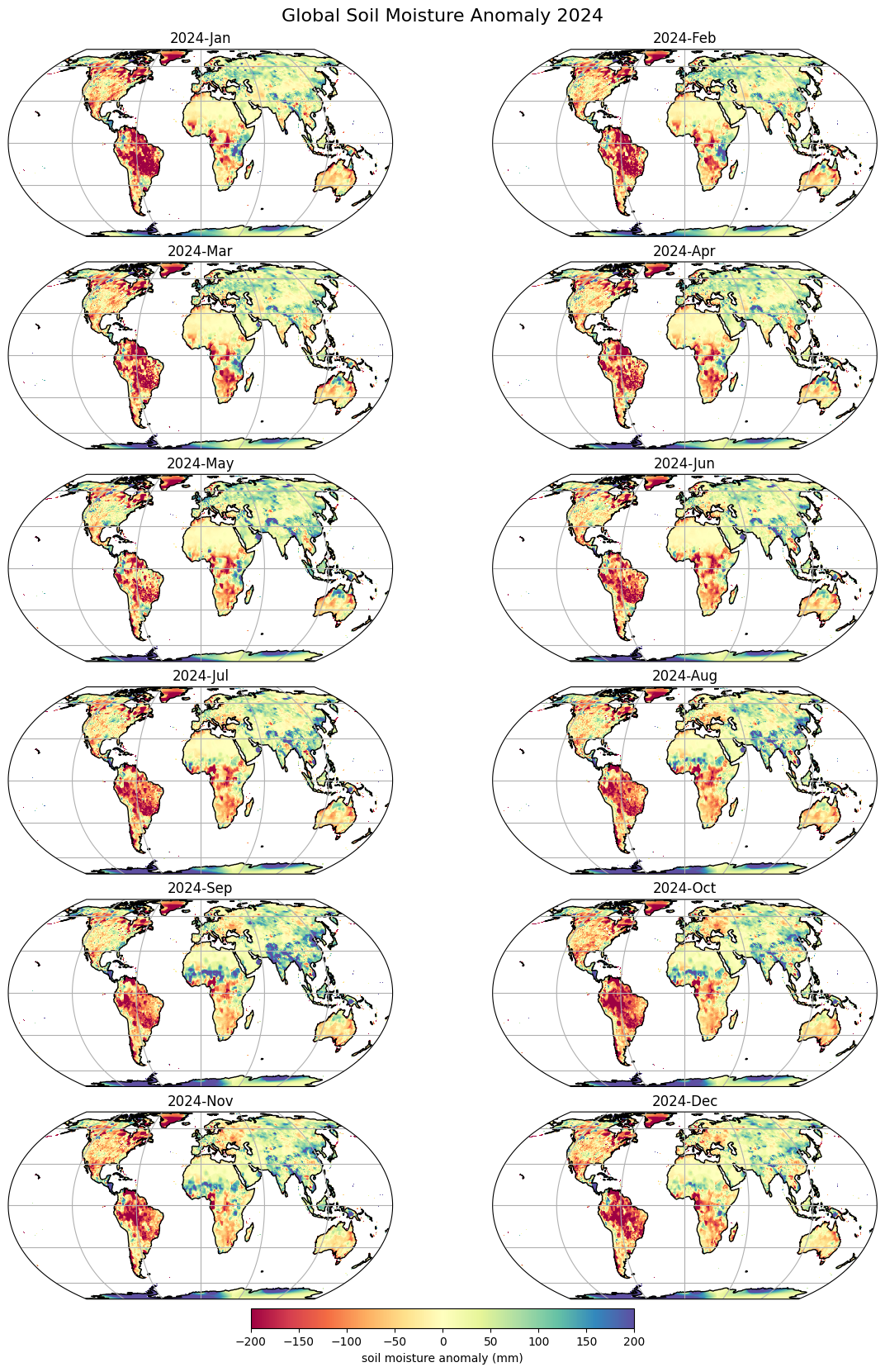
Let’s calculate soil moisture anomaly for 2021 as defined as the departure relative to the 1971–2000 climatology. We group the data by months and calculate monthly means across the entire time perid. We get a DataArray of 12 monthly means. Subtract the mean monthly soil moisture from each month to get the monthly anomalies.
%%time
mean = da.sel(time=slice('1971', '2000')).groupby('time.month').mean('time')
anomaly = da.groupby('time.month') - mean
We now plot the monthly anomaly for each month of 2024.
anomaly2024 = anomaly.sel(time='2024')
We create a layout with 12 subplots (6 rows and 2 columns). The result will be a 2D NumPy array of Axes for each subplot. To easily iterate over each, use use NumPy’s numpy.ndarray.flat attribute to get a flattened 1D array. The enumerate() function will give us the index of the array along with the Axes object from the array.
projection = ccrs.EqualEarth()
fig, axes = plt.subplots(6, 2, sharex=True, sharey=True,
constrained_layout=True,
subplot_kw={'projection': projection})
fig.set_size_inches(11.7, 16.5)
for index, ax in enumerate(axes.flat):
data = anomaly2024.isel(time=index)
im = data.plot(
ax=ax,
transform=ccrs.PlateCarree(), # Data is in PlateCarree
cmap='Spectral',
vmin=-200, vmax=200,
add_colorbar=False, add_labels=False)
title = pd.to_datetime(data.time.values).strftime('%Y-%b')
ax.set_title(title)
ax.set_aspect('equal')
# Add coastlines and gridlines
ax.coastlines()
ax.gridlines()
# Place the colorbar spanning the two bottom axes
fig.colorbar(im, ax=axes[5, :2], shrink=0.4, pad=0.05, location='bottom',
label='soil moisture anomaly (mm)')
fig.suptitle('Global Soil Moisture Anomaly 2024', fontsize=16)
plt.show()
If you want to give feedback or share your experience with this tutorial, please comment below. (requires GitHub account)

