Processing Time-Series with XEE and XArray#
Introduction#
Google Earth Engine (GEE) is a cloud-based platform that has a large public data catalog and computing infrastructure. The XEE extension makes it possible to obtain pre-processed data cube directly from GEE as a XArray Dataset. In this section, we will learn how to process the GEE data using XArray and Dask on local compute infrastructure using the time-series processing capabilities of XArray.
Overview of the Task#
We will obtain a datacube of cloud-masked Sentinel-2 images for a year over a chosen region and apply temporal aggregation to obtain mean monthly NDVI images.
Note: You must have a Google Earth Engine account to complete this section. If you do not have one, follow our guide to sign up.
Output:
2019_01.tif,2019_02.tif, ..: 12 GeoTIFF files of monthly mean NDVI calculated from Sentinel-2 L1C scenes.
Data Credit:
Sentinel-2 Level 1C Scenes: Contains modified Copernicus Sentinel data (2025-02)
Setup and Data Download#
The following blocks of code will install the required packages and download the datasets to your Colab environment.
%%capture
if 'google.colab' in str(get_ipython()):
!pip install --upgrade xee
!pip install rioxarray
import ee
import xarray
import rioxarray as rxr
import matplotlib.pyplot as plt
import pandas as pd
import os
import datetime
import numpy as np
output_folder = 'output'
if not os.path.exists(output_folder):
os.mkdir(output_folder)
We initialize Google Earth Engine API.You must have a Google Cloud Project associated with your GEE account. Replace the cloud_project with your own project from Google Cloud Console. Here we are using the High Volume Endpoint which is recommended when working with XEE.
cloud_project = 'spatialthoughts'
try:
ee.Initialize(project=cloud_project, opt_url='https://earthengine-highvolume.googleapis.com')
except:
ee.Authenticate()
ee.Initialize(project=cloud_project, opt_url='https://earthengine-highvolume.googleapis.com')
Data Pre-Processing#
We start with the Sentinel-2 collection, apply a cloud mask and compute NDVI using the Google Earth Engine API.
s2 = ee.ImageCollection('COPERNICUS/S2_HARMONIZED')
geometry = ee.Geometry.Polygon([[
[82.60642647743225, 27.16350437805251],
[82.60984897613525, 27.1618529901377],
[82.61088967323303, 27.163695288375266],
[82.60757446289062, 27.16517483230927]
]])
filtered = s2 \
.filter(ee.Filter.date('2019-01-01', '2020-01-01')) \
.filter(ee.Filter.lt('CLOUDY_PIXEL_PERCENTAGE', 30)) \
.filter(ee.Filter.bounds(geometry))
# Write a function for Cloud masking
def maskS2clouds(image):
qa = image.select('QA60')
cloudBitMask = 1 << 10
cirrusBitMask = 1 << 11
mask = qa.bitwiseAnd(cloudBitMask).eq(0).And(
qa.bitwiseAnd(cirrusBitMask).eq(0))
return image.updateMask(mask).multiply(0.0001) \
.select('B.*') \
.copyProperties(image, ['system:time_start'])
filtered = filtered.map(maskS2clouds)
# Write a function that computes NDVI for an image and adds it as a band
def addNDVI(image):
ndvi = image.normalizedDifference(['B8', 'B4']).rename('ndvi')
return image.addBands(ndvi)
# Map the function over the collection
withNdvi = filtered.map(addNDVI)
Now we have an ImageCollection that we want to get it as a XArray Dataset. We define the region of interest and extract the ImageCollection using the ‘ee’ engine.
scale in XEE behaves differently than in GEE API. The value of the scale is in the units of the CRS. So to get the data at 10 meters resolution, we must also specify a CRS that has the unit of meters.
ds = xarray.open_dataset(
withNdvi.select('ndvi'),
engine='ee',
crs='EPSG:3857',
scale=10,
geometry=geometry,
)
ds
Since we want to work with coordinates in latitude and longitude, we reproject the raster to EPSG:4326. This requires renaming and reordering the dimentions to those expected by rioxarray.
ds_reprojected = ds\
.rename({'Y': 'y', 'X': 'x'}) \
.transpose('time', 'y', 'x') \
.rio.reproject('EPSG:4326')
ds_reprojected
Select the ndvi variable. Split the datacube into ‘chunks’ to allow parallel processing using Dask.
original_time_series = ds_reprojected.ndvi.chunk('auto')
original_time_series
Aggregating the Time-Series#
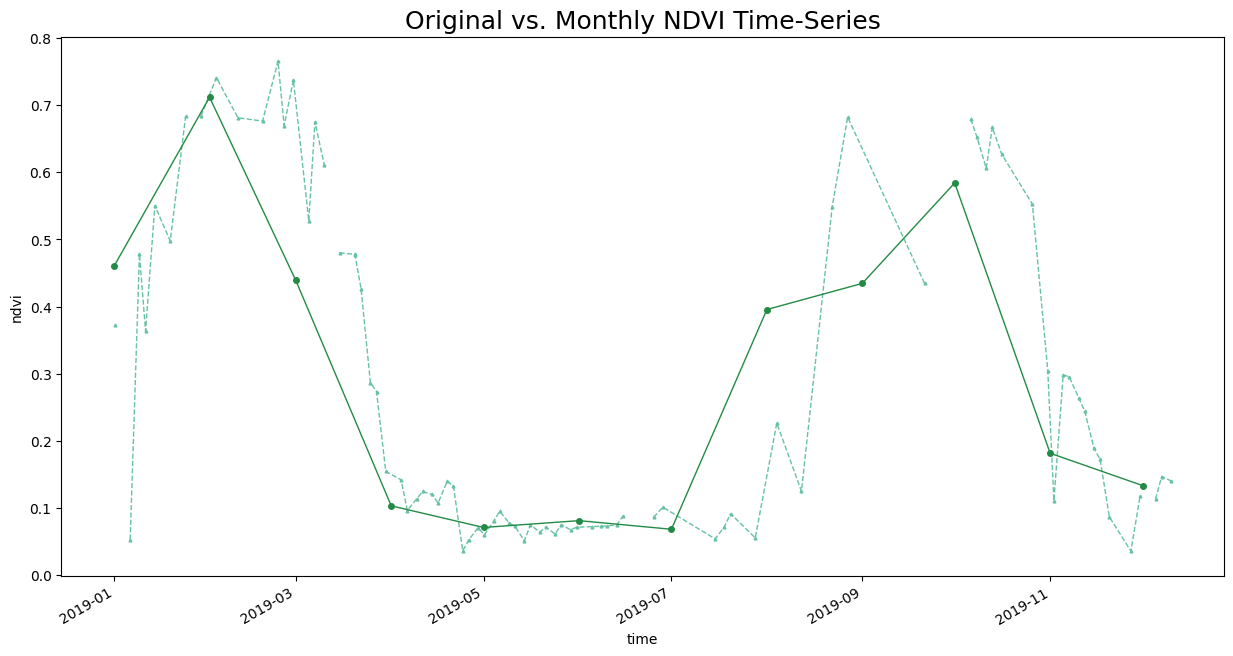
We have a irregular time-series with a lot of noise. Let’s create a monthly NDVI time-series by computing the monthly average NDVI. We can use XArray’s resample() method to aggregate the time-series by Months and coompute the mean. We specify time as MS to indicate aggregating by month start and label as left to indicate we want the start of the month as our series labels.
Reference Pandas Offset Aliases
time_series_aggregated = original_time_series.resample(time='MS', label='left').mean()
time_series_aggregated
Plot and Extract the Time-Series at a Single Location
original_ts = original_time_series.interp(x=82.607376, y=27.164335).compute()
aggregated_ts = time_series_aggregated.interp(x=82.607376, y=27.164335).compute()
fig, ax = plt.subplots(1, 1)
fig.set_size_inches(15, 7)
original_ts.plot.line(
ax=ax, x='time',
marker='^', color='#66c2a4', linestyle='--', linewidth=1, markersize=2)
aggregated_ts.plot.line(
ax=ax, x='time',
marker='o', color='#238b45', linestyle='-', linewidth=1, markersize=4)
ax.set_title('Original vs. Monthly NDVI Time-Series', size = 18)
plt.show()
Downloading Time-Series Images#
Save the processed monthly NDVI images using rioxarray as GeoTIFF files.
for time in time_series_aggregated.time.values:
image = time_series_aggregated.sel(time=time)
date = np.datetime_as_string(time, unit='M')
output_file = f'{date}.tif'
output_path = os.path.join(output_folder, output_file)
image.rio.to_raster(output_path, driver='COG')
print(f'Created file at {output_path}')
If you want to give feedback or share your experience with this tutorial, please comment below. (requires GitHub account)

